Postdoctoral Fellows
Current
B2/L2 - Sea Side - 2217-WS01
Ph.D. research: [Mycobacterial phylogenomics and stress response metrics]
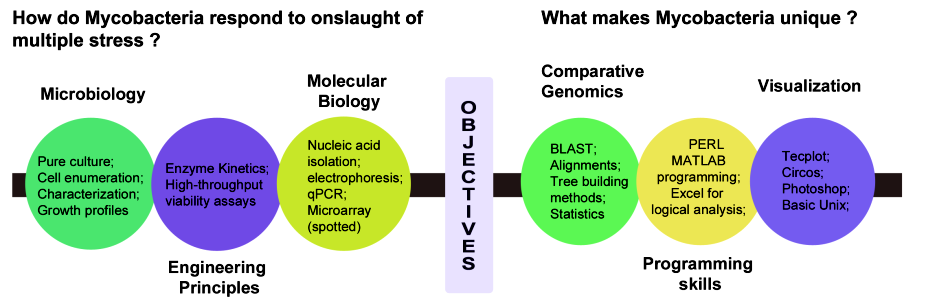
My passion for research at a higher level made me resign my job as a member of the faculty at the biological sciences division (BITS Pilani, India) to pursue Ph.D. at IIT Bombay, India (details in C.V). The objective of my thesis was related to long-lasting puzzle concerning Mycobacterium physiology using M.smegmatis as the model system. I employed computational and experimental approaches to address the questions we asked (shown in graphics below).
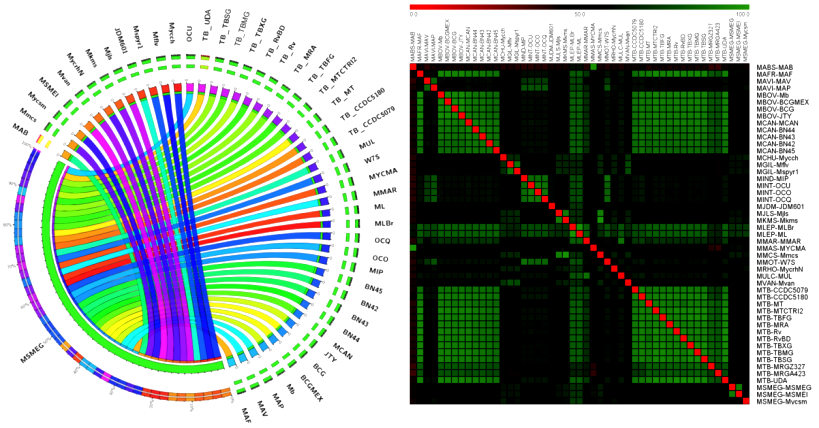
First, I used a comparative genomics approach to understand the unique features of pathogenic and non-pathogenic mycobacteria. My results established the relation and distinction between two groups regarding, the uniqueness of their genome, number of shared orthologs, global genome rearrangement patterns and the genes that are exclusive to pathogens (Interesting read).
Second, I performed experimental work to quantify the response of M. smegmatis (Wild-type and sigma factor mutants) to multiple stresses, both at phenotypic and gene level. I chose four stressors including exposure to various pH; heat shock, cold shock, and starvation. We employed a tri-parametric approach to quantify stress response namely, measurement of metabolic activity, doubling ability and adaptability. I developed a high-throughput-viability assay to measure the phenotypic response via metabolic activity. For the other two parameters, I used colony count and growth profiles respectively. Further, I analyzed the gene expression using quantitative Real-time PCR for selected multi-stress conditions which was lethal to cells.
Results of my research work are important in the field of Mycobacterial research. Specifically, comparative phylogenomics revealed that the pathogenicity of each mycobacterium is unique and mycobacterial pathogens underwent more rearrangements within core genes compared to non-pathogens. On the other hand, multi stress studies, a first of its kind in Mycobacteria showed the robustness of M. smegmatis.
First Post-doc research: [Fungal transcriptomics and Phylogenomics]
My inclination towards academia led me to seek a postdoctoral fellowship. For my first postdoc, I wanted to contribute and further hone my expertise by working on a project that had scope for phylogenomics and transcriptomics. Fortunately, I landed as a post-doctoral researcher at Fungal evolution and genomics laboratory, under the guidance of Prof. Laszlo Nagy, BRC, Szeged, Hungary. I worked on several projects with a high point being my hard work in a project on “forest killer” fungi named Armillariaostoyae (Story behind the paper !). Our research was on international news and published in Nature Ecology and Evolution (Interesting read), given the cruciality of the problem (snapshot is shown below).
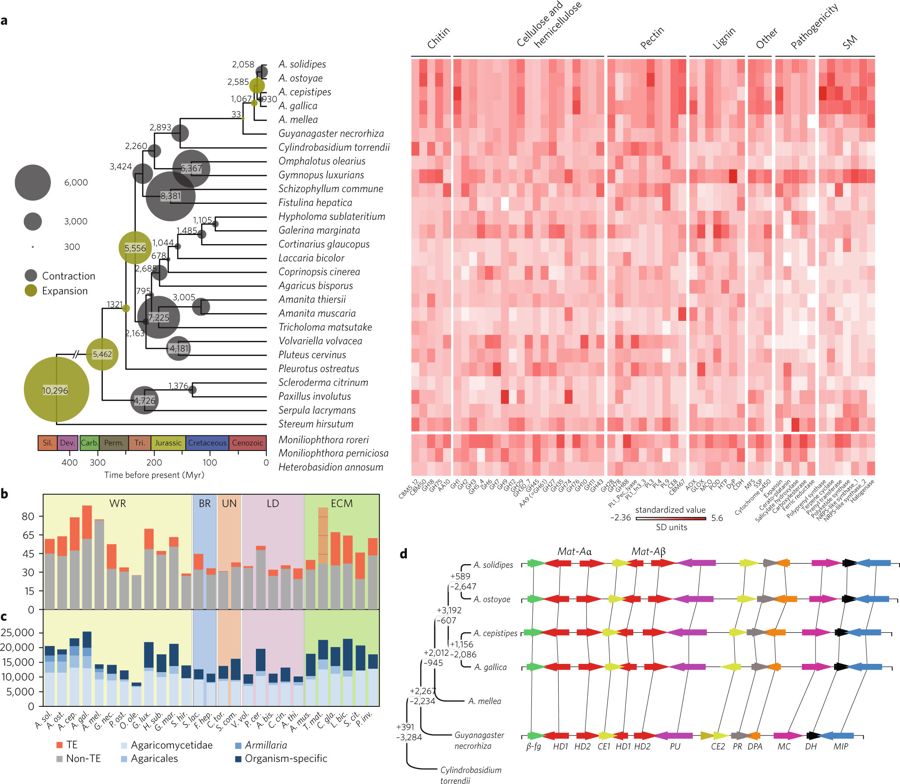
Briefly, I aided RNA sequencing for the invasive and reproductive developmental stages of A. ostoyae including rhizomorphs and compared its whole genome with 22 related fungi. Our results revealed a significant genome expansion in Armillaria, affecting several pathogenicity-related genes, lignocellulose-degrading enzymes and lineage-specific genes expressed during rhizomorph development. Rhizomorphs express an evolutionarily young transcriptome that shares features with the transcriptomes of both fruiting bodies and vegetative mycelia. Several genes showed concomitant upregulation in rhizomorphs and fruiting bodies and shared cis-regulatory signatures in their promoters, providing genetic and regulatory insights into complex multicellularity in fungi. Our results suggested that the evolution of the unique dispersal and pathogenicity mechanisms of Armillaria might have drawn upon ancestral genetic toolkits for wood-decay, morphogenesis, and complex multicellularity.
Current research at Aranda Lab: [Transcriptomics, Genomics, and Bioinformatics]
My enthusiasm towards research at the molecular level grew consistently over time leading to the need for a more profound understanding of NGS technology and related skills. This quest led me to pursue a position in Prof. Aranda’s coral symbiomics laboratory, whose research objectives and aptitude I found aligning with my research interest. Here at coral symbiomics lab, I am exploring and working in projects that involve the study of molecular mechanism of coral symbiosis using sea anemone as the model organism. The projects involve a high quotient of experimental work and bioinformatics viz. genome sequencing, transcriptome sequencing, IsoSeq and gene network analysis (but not limited to).
Gladly, I secured two-year funding through “Competitive Research Grant (CRG)” for the year 2018, to identify genetic drivers of thermal tolerance plasticity in the coral model Exaiptasia. Successful completion of our proposal will lead to expanding the community knowledge on the coral model, identification of new candidate genes for further functional genomic studies and reconstruction of a high-resolution genetic footprint. Positively, we will be able to extrapolate the conclusions to corals and help in the advancement of coral research for their better future.
Always up for scientific and productive discussions! Feel free to write to me: arun.nagarajan@kaust.edu.sa or arunprasanna83@gmail.com
● Red Sea Research Center (RSRC)
● Biological and Environmental Science and Engineering Division (BESE)